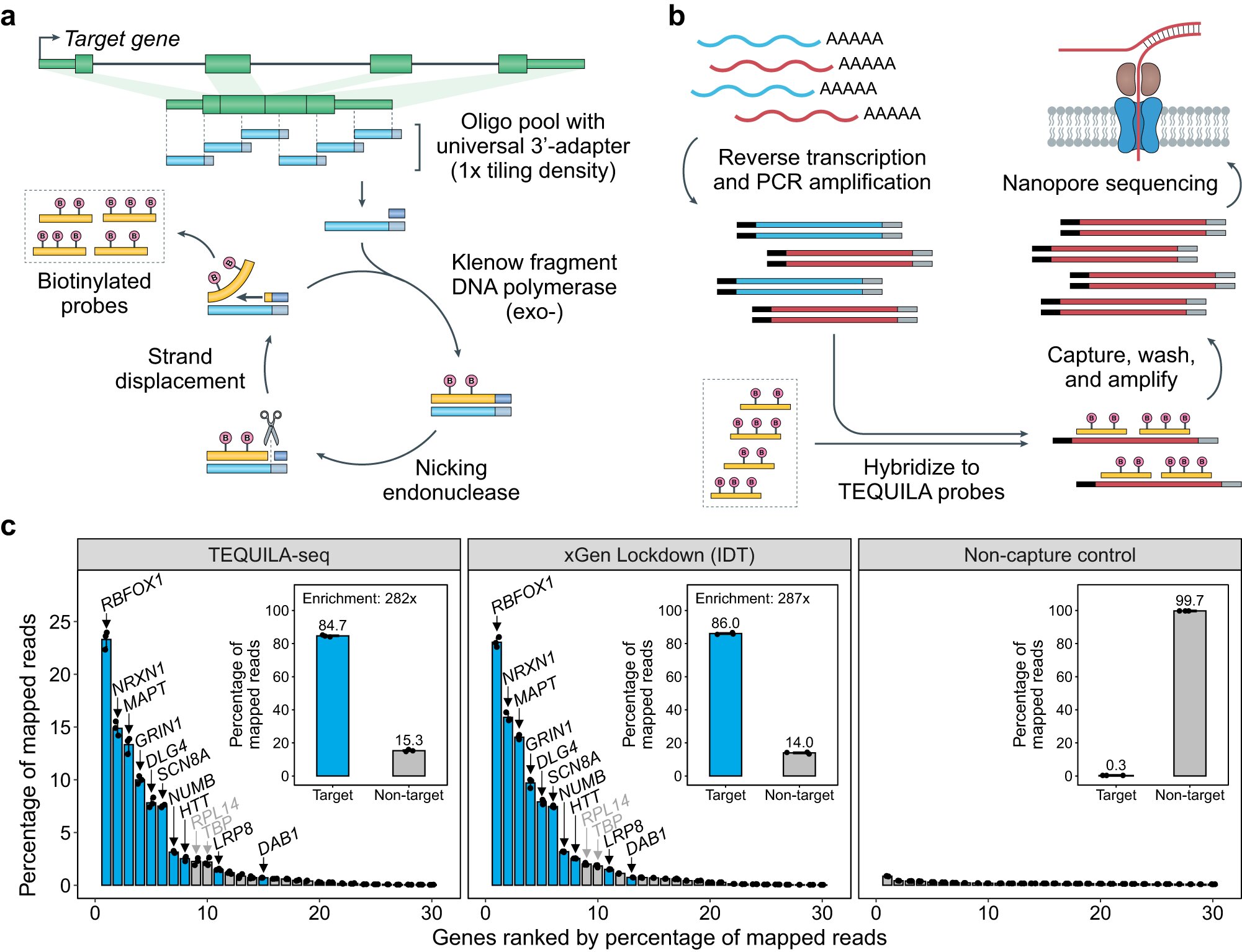
In a development that could accelerate the discovery of new diagnostics and treatments, researchers at Children’s Hospital of Philadelphia (CHOP) have developed a versatile and low-cost technology for targeted sequencing of full-length RNA molecules. The technology, called TEQUILA-seq, is highly cost-effective compared to commercially available solutions for targeted RNA sequencing and can be adapted for different research and clinical purposes. The details are described in a paper in Nature Communications.
On the journey from gene to protein, an RNA molecule can be cut and joined in different ways before being translated into a protein. This process is known as alternative splicing, and it allows a single gene to encode several different proteins. Although alternative splicing occurs in many biological processes, it can be dysregulated in diseases like cancer, leading to pathogenic RNA molecules. To understand how alternative splicing might lead to disease, researchers need to have accurate accounting of all the RNA molecules (known as “transcript isoforms”) that emanate from a single gene.
One way of doing so is using “long-read” RNA sequencing platforms, which sequence RNA molecules over 10,000 bases in length end-to-end, capturing the entirety of the transcript isoforms. However, these long-read platforms have modest sequencing yield, which has hampered their widespread adoption, especially in the clinical setting, as generating long-read RNA sequencing data at clinically informative depth could be prohibitively expensive. Targeted sequencing, which involves enriching specific nucleic acid sequences of interest prior to sequencing, is a useful strategy that can substantially enhance coverage of predefined targets, but the cost and complexity of target capture have been barriers to wider use.
“Targeted long-read RNA sequencing is a powerful strategy for elucidating the RNA repertoire for any predefined set of genes. However, existing technologies for targeted sequencing of full-length RNA molecules are either expensive or difficult to set up, putting them out of reach for many labs,” said co-senior author Lan Lin, Ph.D., Assistant Professor of Pathology and Laboratory Medicine and a member of the Raymond G. Perelman Center for Cellular and Molecular Therapeutics at CHOP. “TEQUILA-seq solves that problem by being both inexpensive and easy to use. The technology can be adapted by users for different purposes, and researchers can choose which genes they want to sequence and make the reagents for target capture in their own labs. This has the potential to accelerate discovery of new diagnostic and therapeutic solutions for a wide range of diseases.”
One method that allows for targeted sequencing is called hybridization capture-based enrichment, which uses short pieces of nucleic acids called oligonucleotides as capture probes. These oligonucleotides (often simply referred to as “oligos”) are tagged with biotin molecules and designed to hybridize to their targets based on nucleic acid sequence complementarity, which allows for easy capture and isolation of their target sequences from a biological sample. However, although hybridization capture-based enrichment is an efficient method for targeted sequencing, commercially synthesized biotinylated capture probes are expensive and can only be used for a limited number of reactions, making the per-sample cost high for each capture reaction.
2023-08-15 23:24:02
Original from phys.org rnrn