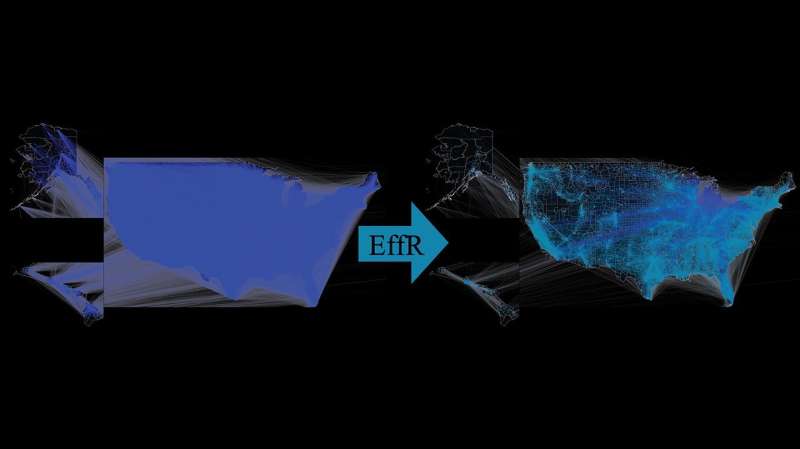
Fig 1. Sparsification of the U.S. mobility community. On the left is the unique community with about 26 million edges. On the proper, a sparsified community primarily based on efficient resistance sampling. Credit: PLOS Computational Biology (2022). DOI: 10.1371/journal.pcbi.1010650
Simulations that assist decide how a large-scale pandemic will unfold can take weeks and even months to run. A latest examine in PLOS Computational Biology presents a brand new method to epidemic modeling that would drastically pace up the method.
The examine makes use of sparsification, a way from graph principle and laptop science, to determine which hyperlinks in a community are crucial for the unfold of illness.
By specializing in important hyperlinks, the authors discovered they might cut back the computation time for simulating the unfold of ailments by extremely advanced social networks by 90% or extra.
“Epidemic simulations require substantial computational sources and time to run, which suggests your outcomes could be outdated by the point you’re able to publish,” says lead creator Alexander Mercier, a former undergraduate analysis fellow at SFI and now a Ph.D. scholar on the Harvard T.H. Chan School of Public Health. “Our analysis might in the end allow us to make use of extra advanced fashions and bigger information units whereas nonetheless appearing on an inexpensive timescale when simulating the unfold of pandemics reminiscent of COVID-19.”
For the examine, Mercier, with SFI researchers Samuel Scarpino and Cristopher Moore, used information from the U.S. Census Bureau to develop a mobility community describing how folks throughout the nation commute.
Then, they utilized a number of completely different sparsification strategies to see if they might cut back the community’s density whereas retaining the general dynamics of a illness spreading throughout the community.
The most profitable sparsification…
2023-01-05 07:14:31 New method to epidemic modeling might pace up pandemic simulations
Post from phys.org