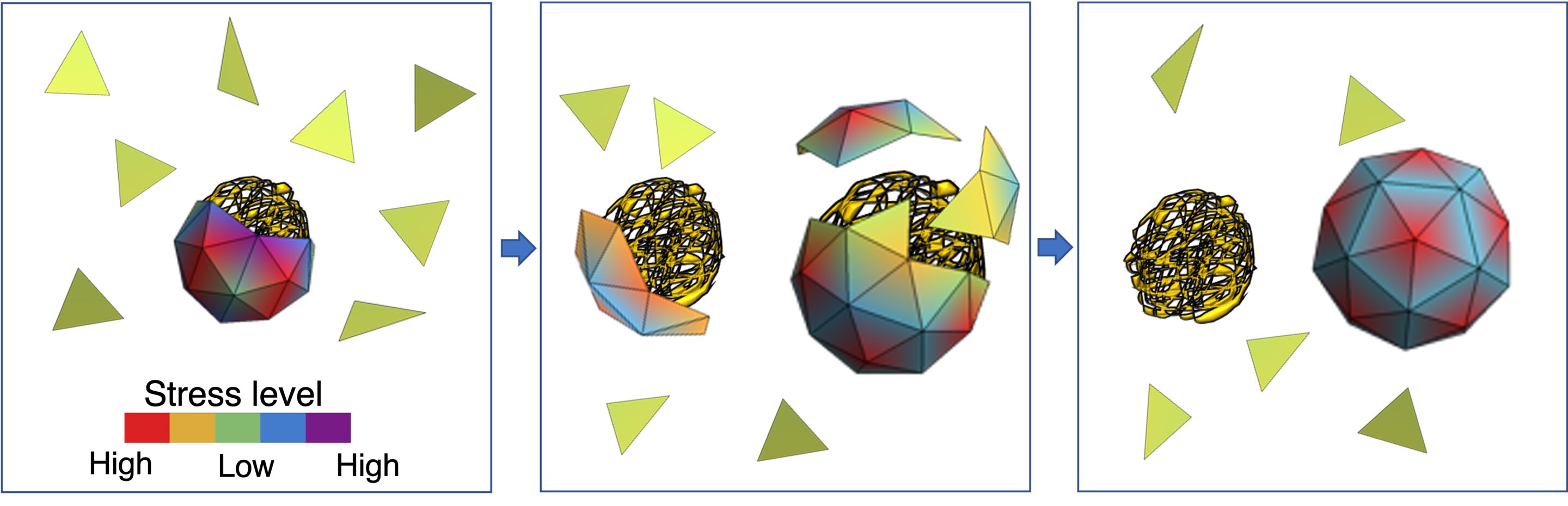
From left to proper: The inexperienced triangles mimic capsid proteins. In the absence of a viral genome, they assemble round smaller segments of RNA. However, the shell is careworn and might be simply damaged into items. The coloration bar reveals the totally different ranges of stress. Once the viral RNA is offered, the capsid proteins begin to go away non-viral RNAs and assemble across the viral genome. This might be accomplished comparatively simply due to the excessive stress degree of capsid proteins within the smaller shell. The proper determine reveals capsid proteins assembled across the native RNA, forming a steady icosahedral shell. Credit: Zandi lab, UC Riverside.
Each easy RNA virus has a genome, its “native RNA.” This genome dictates how the virus replicates in cells to finally trigger illness. The genome additionally has the code for making a capsid, the protein shell of a virus that encapsulates the genome and protects it like a nanocontainer.
A group led by Roya Zandi, a professor of physics and astronomy on the University of California, Riverside, has developed a concept and carried out a sequence of simulations which will assist clarify how a virus finds its native genome and the way capsids type round it and never round different RNAs within the cell.
“A greater understanding of how capsids type is of significant significance to materials scientists and a vital step within the design of engineered nano-shells that might function autos for delivering medication to particular targets within the physique,” Zandi mentioned.
The researchers’ work, printed in ACS Nano, reveals that the interaction of the mechanical properties of proteins, the dimensions of the genome, and the energy of the interplay between the genome and capsid proteins can considerably modify the symmetry, construction, and stability of the capsid.
When a virus enters a cell, the capsid breaks open to launch the genome, which then makes use of the cell’s reproductive equipment to copy. The newly fashioned genomes start to accumulate their capsids, a course of primarily pushed by the enticing electrostatic interplay between the optimistic fees on capsid proteins and the destructive fees on the genomes. But how the virus selects and packages its native RNA contained in the crowded atmosphere of a bunch cell cytoplasm within the presence of many nonviral RNA and different polymers has remained a thriller.
The simulations run by Zandi’s group present that capsid proteins might, in concept, decide any nonviral genome to encapsulate. But the viral genome, she mentioned, is greatest suited to capsid proteins to type a shell round on account of an interaction of energies on the molecular degree.
“The stress distribution of the capsid proteins is decrease when the capsids encapsulate their very own genome, the one for which they have been coded,” Zandi mentioned. “The vitality of the entire system is decrease. While smaller nonviral RNAs can be found within the cell in lots, the capsid proteins are inclined towards forming a shell round a viral RNA as a result of the ensuing soccer ball-like shell has a decrease stress distribution.”
Zandi mentioned the work lays out a scientific comparability of concept and experiments, which is able to permit a greater understanding of the position of RNA within the capsid meeting pathway, stability, and construction.
“A deeper understanding of the position of the genome in virus meeting mechanisms might result in design ideas for different antivirals,” she mentioned.
The new work is an early step in understanding viral meeting. The course of just isn’t properly understood as a result of viruses measure in nanometers and the meeting happens in milliseconds.
“Theoretical work and simulations are needed to grasp how a virus grows,” Zandi mentioned.
Zandi was joined within the analysis by graduate pupil Sanaz Panahandeh at UCR; Siyu Li at Songshan Lake Materials Laboratory in China; and Bogdan Dragnea at Indiana University, Bloomington. Li is a former graduate pupil at UCR.
How a virus varieties its symmetric shells
More info:
Sanaz Panahandeh et al, Virus Assembly Pathways Inside a Host Cell, ACS Nano (2022). DOI: 10.1021/acsnano.1c06335
Provided by
University of California – Riverside
Citation:
Simulations on how a virus packages its genetic materials might assist design nanocontainers utilized in drug supply (2022, March 9)
retrieved 9 March 2022
from https://phys.org/information/2022-03-simulations-virus-packages-genetic-material.html
This doc is topic to copyright. Apart from any truthful dealing for the aim of personal research or analysis, no
half could also be reproduced with out the written permission. The content material is supplied for info functions solely.