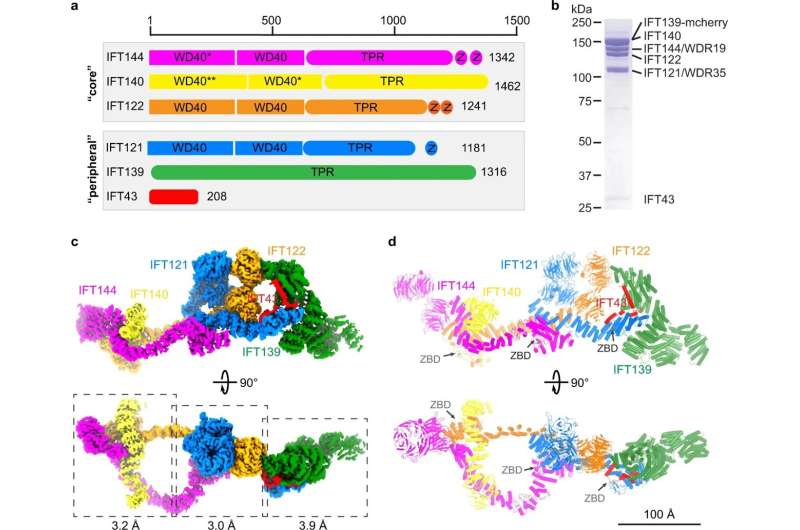
Structure of the IFT-A complex. a Domain architecture of the IFT-A subunits. The “core” and “peripheral” subcomplexes are indicated in gray boxes. WD40, TPR and Z stand for WD40, TPR-like and zinc-binding domains, respectively. * indicates that the domain is resolved at low resolution but we could use AF2 model to do rigid body docking. ** indicates that the domain is not resolved due to flexibility. b SDS-PAGE of the purified IFT-A complex. IFT139 is tagged with mCherry. c Cryo-EM density of the IFT-A complex. IFT144, IFT140, IFT139, IFT122, IFT121 and IFT43 are colored in magenta, yellow, green, orange, blue and red, respectively. The same color codes are used in the manuscript unless otherwise noted. The local refinement resolutions are indicated. d Structural model of the IFT-A complex shown as cartoons. The ZBDs are indicated by gray arrows. Credit: Cell Research (2023). DOI: 10.1038/s41422-023-00778-3
St. Jude Children’s Research Hospital scientists solved the 3D structure of a major protein complex in cilia, signaling appendages found on cells. The structure was captured at the highest resolution to date. The work serves as a foundation to study diseases of the brain, kidney, skeleton and eyes that are known to involve cilia but were difficult to investigate. The findings were published today in Cell Research.
Cilia are hair-like projections found on nearly all mammalian cells that control many signaling processes. The cilium is a type of thin tube that lacks the machinery to synthesize the proteins needed for cell signaling. Therefore, the signaling molecules inside cilia must be brought over from other areas in the cell. Intraflagellar transport complexes, called IFT-A and IFT-B, serve as trains that bring proteins to and from cilia.
For over a decade, scientists have attempted to understand the structure of the IFT-A and -B complexes. The St. Jude scientists determined the structure of IFT-A to an overall resolution around 3-4 Angstroms, which allows us to visualize these complexes in near-atomic detail.
“Now that we have the high-resolution structure of this ciliary complex, we can map mutations known to cause diseases and then design clinical interventions,” said co-corresponding author Ji Sun, Ph.D., St. Jude Department of Structural Biology. “We can tell at the atomic level tell how these trains assemble with each other to form very elegant structures in the cilia. We can also use that knowledge to understand how disease mutations disrupt that structure.”
Their work reveals new details that were never clear enough to understand in previous attempts. For example, they revealed previously unknown zinc-binding sites in IFT-A. Zinc-binding sites are important to a type of protein domain called zinc fingers. Zinc fingers are critical for certain protein-protein interactions, which explained some poorly understood connections within the train complex.
“It was pretty exciting to see the zinc fingers because no one had seen or even predicted zinc-binding sites in IFT-A,” Sun said. “Our study was able to reveal that with confidence. We could say, ‘Hey, there’s zinc, and it is important for protein-protein interaction, which might also facilitate train assembly.’ We never would have figured that out without our high-resolution structural information.”
2023-02-13 11:40:45
Article from phys.org
Molecular motors are proteins that are able to move along biological filaments and are key components of a variety of cellular functions, including cilia movement. Cilia are microscopic, hairlike structures found on the surface of certain cells and are essential for a variety of functions, including maintaining body temperature and moving mucus. An international team of researchers has recently developed a novel approach that allows for the tracking of individual molecular motors as they power cilia movement.
The researchers used optical tweezers microscopy to track the motion of individual molecular motors from a variety of cilia-producing cells. This allowed them to observe both the motion of single cilia, as well as the speed and force generated by individual molecular motors.
The data acquired during this study revealed a surprising degree of variability in the speed and force generated by individual molecular motors. The researchers hypothesized that this variation may be due to differences in the structure of the filament along which these molecular motors travel, or to variations in the number and type of proteins associated with the motors.
The researchers further observed that the speed of the molecular motors increases when the cilia are exposed to increased concentrations of a particular protein. This suggests that the movement of the motors is directly correlated with cilia activity, and that higher concentrations of a particular protein may lead to faster cilia movement.
Overall, this study demonstrates the potential of optical tweezers microscopy to track the motion of individual molecular motors and provides insight into the mechanisms underlying cilia movement. The results of this study could lead to a better understanding of the roles of proteins associated with cilia in regulating various cellular functions, as well as further development of treatments for cilia-related illnesses.
