New bacterial pathogen pipeline makes entire genome sequencing accessible for public well being scientists worldwide. Credit: Earlham Institute
A worldwide consortium of scientists, led by the Earlham Institute and the University of Liverpool within the UK, mark a major milestone in equipping researchers in low- and middle-income nations (LMICs) with low-cost and accessible strategies for sequencing massive collections of bacterial pathogens—at a price of lower than $10USD per genome.
At a time when international genomic surveillance of COVID-19 coronavirus has been within the highlight, the power of nations to contribute by way of low-cost and fast entire genome sequencing (WGS) has grow to be more and more necessary. The strategies revealed in Genome Biology may be utilized to massive collections of bacterial pathogens and can strengthen international analysis collaborations to sort out future pandemics.
Over the previous decade, WGS has revolutionized understanding of microbial epidemics. WGS information can be utilized for surveillance, practical genomics and the exploration of pathogen evolution, prompting each public well being and analysis scientists to undertake genome-based approaches.
The genome sequencing of hundreds of microorganisms has remained costly—largely as a consequence of prices related to pattern transportation and the development of DNA libraries—whereas the necessity to genome sequence collections of key pathogens has grown considerably lately.
Until now, large-scale bacterial genome initiatives may solely be carried out in a handful of sequencing facilities all over the world. With this examine, the workforce of scientists have managed to make this know-how accessible to laboratories worldwide.
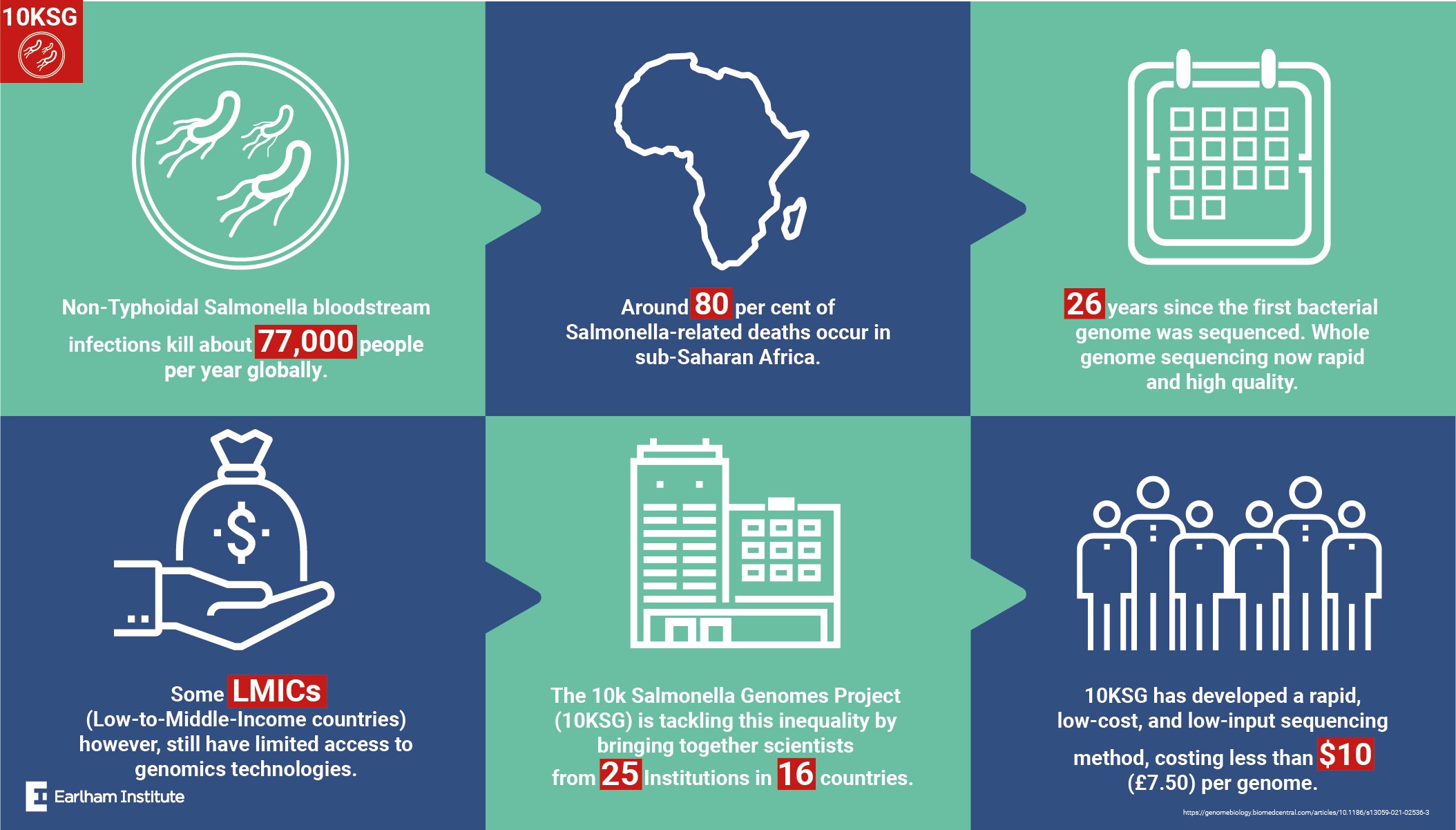
“It has been 26 years because the first bacterial genome was sequenced, and it’s now doable to sequence bacterial isolates at scale. However, entry to this game-changing know-how for scientists in low- and middle-income nations has remained restricted, stated examine writer and Director of the Earlham Institute Prof Neil Hall.
“The must ‘democratise’ the sphere of pathogen genomic evaluation prompted us to develop a brand new technique to sequence hundreds of bacterial isolates with collaborators primarily based in lots of economically-challenged nations.”
10k Salmonella strains
Focusing on the organism Salmonella enterica, a pathogen with a world significance that causes an infection and lethal illness, this large-scale genomic sequencing initiative was led by the worldwide 10,000 Salmonella genomes analysis consortium (10KSG) with scientists from 16 nations.
The goals of 10KSG are to make genomic information extra accessible to low and center revenue nations, particularly as a result of mortality charges for Salmonella in sub-Saharan Africa are exceptionally excessive. Understanding the genetic make-up of serious collections of such micro organism strains was crucial, and the venture sequenced and analyzed 10,000 Salmonella genomes from Africa and Latin America.
The researcher’s modern WGS strategy aimed to streamline the large-scale acquisition and genome sequencing of micro organism, and amassed the genetic materials of greater than 10,400 scientific and environmental bacterial isolates from LMICs in below a yr.
The pattern logistics pipeline, developed by the University of Liverpool, was optimized by transport the heat-inactivated bacterial isolates as ‘thermolysates’ in ambient situations from the world over to the UK. Subsequently, isolates have been sequenced on the Earlham Institute utilizing the distinctive LITE protocol—a low price, low enter automated methodology for fast genome sequencing. In complete, the gene library building and DNA sequencing bioinformatic evaluation was carried out with a complete reagent price of lower than USD$10 (round £7.50GBP) per genome.
Prof of Microbial Pathogenesis and examine writer Jay Hinton from the University of Liverpool, stated: “One of essentially the most important challenges dealing with public well being researchers in LMI nations is entry to state-of-the-art know-how. For a mix of logistical and financial causes, the areas related to the best burden of extreme bacterial illness haven’t benefited from widespread availability of WGS. The 10,000 Salmonella genomes venture was designed to start to handle this inequality.”
Dr. Blanca Perez Sepulveda, Postdoctoral Research Associate and examine writer from the University of Liverpool, who led the worldwide pattern assortment, optimisation and evaluation, added: “The adoption of large-scale genome sequencing and evaluation of bacterial pathogens will likely be an infinite asset to public well being and surveillance in LMI nations. Here, we now have established an environment friendly and comparatively cheap pipeline for the worldwide assortment and sequencing of bacterial genomes.”
Deadly micro organism
Non-typhoidal Salmonella (NTS) have been extensively related to enterocolitis in people, a zoonotic illness that’s linked to the industrialisation of meals manufacturing. Due to the size of human instances of enterocolitis and issues associated to meals security, extra genome sequences have been generated for Salmonella than some other genus.
In current years, new lineages of NTS serovars Typhimurium and Enteritidis have been acknowledged as widespread causes of invasive bloodstream infections (iNTS illness), accountable for about 77,000 deaths per yr worldwide.
Approximately 80 % of deaths as a consequence of iNTS illness happen in sub-Saharan Africa. The new Salmonella lineages accountable for bloodstream infections may be recognized by genomics, as a consequence of gene degradation, altered prophage repertoires and novel multidrug resistant plasmids.
Prof Neil Hall, added: “The variety of publicly-available sequenced Salmonella genomes reached 350,000 in 2021 and can be found from a number of on-line repositories. However, restricted genome-based surveillance of Salmonella infections has been carried out in LMI nations, and the prevailing dataset didn’t precisely symbolize the Salmonella pathogens which might be at the moment inflicting illness the world over.”
Global pipeline
Dr. Darren Heavens, Postdoctoral Scientist on the Earlham Institute, who developed the whole-genome sequencing pipeline, stated: “We noticed the necessity to simplify and increase genome-based surveillance of salmonellae from Africa and different components of the world, involving isolates related to invasive illness and gastroenteritis in people, and lengthening to micro organism derived from animals and the surroundings.
“Our pipeline represents a cheap and strong software for producing bacterial genomic information from LMI nations, to permit investigation of the epidemiology, drug resistance and virulence elements of isolates.”
Development of the worldwide 10KSG consortium that concerned collaborators from 25 establishments, analysis and reference laboratories throughout 16 nations. Members of the 10KSG offered entry to 10,419 bacterial isolates sourced from 51 LMICs and areas—overlaying seven bacterial genera: Acinetobacter, Enterobacter, Klebsiella, Pseudomonas, Shigella, and Staphylococcus—coordinating the pattern assortment and transport of supplies to be sequenced within the UK.
“Limited funding sources led us to design a genomic strategy that ensured correct pattern monitoring and captured complete metadata for particular person bacterial isolates whereas maintaining prices to a minimal for the Consortium,” stated Prof Hall. “The pipeline streamlined the large-scale assortment and sequencing of samples from LMICs. A key driver was to facilitate entry to WGS and permit a worldwide collaborative effort to generate a remarkably informative and strong set of genomic information.”
The paper “An accessible, environment friendly and international strategy for the large-scale sequencing of bacterial genomes” is revealed in Genome Biology.
Genome scientists use UK Salmonella instances to make clear African epidemic
More data:
An accessible, environment friendly and international strategy for the large-scale sequencing of bacterial genomes, Genome Biology (2021). DOI: 10.1186/s13059-021-02536-3
Provided by
Earlham Institute
Citation:
Affordable genome sequencing for pathogen evaluation to assist sort out international epidemics (2021, December 20)
retrieved 21 December 2021
from https://phys.org/information/2021-12-genome-sequencing-pathogen-analysis-tackle.html
This doc is topic to copyright. Apart from any truthful dealing for the aim of personal examine or analysis, no
half could also be reproduced with out the written permission. The content material is offered for data functions solely.